The Power Spectrum of Survey Data¶
In this notebook, we explore the functionality of the
ConvolvedFFTPower algorithm, which computes the power spectrum
multipoles \(P_\ell(k)\) for data from a survey that includes
non-trivial selection effects. The output of the algorithm is the true
multipoles of the data, convoled with the window function of the survey.
The input data for this algorithm is assumed to be from an observational survey, with the position coordinates specified in terms of right ascension, declination, and redshift.
Note
The data used in this notebook is not realistic – rather, we choose the
simplicity of generating mock data to help users get up and running
quickly. Although the end results are not cosmologically interesting, we
use the mock data to help illustrate the various steps necessary to use
the ConvolvedFFTPower algorithm properly.
In [1]:
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
In [2]:
from nbodykit.lab import *
from nbodykit import setup_logging, style
from scipy.interpolate import InterpolatedUnivariateSpline
import matplotlib.pyplot as plt
plt.style.use(style.notebook)
In [3]:
setup_logging()
Initalizing Mock Data¶
We start by generating mock catalogs to mimic the “data” and “randoms”
catalogs needed by the ConvolvedFFTPower algorithm. Here, the “data”
catalog usually gives the information about the galaxy objects, and the
“randoms” catalog is a catalog of synthetic objects without any
cosmological clustering signal. The “randoms” usually have a higher
number density than the “data” and are a Monte Carlo representation of
the survey volume.
In this notebook, we construct our fake “data” and “randoms” catalogs simply by generating uniformly distributed right ascension and declination values within a region of the sky, with redshifts drawn from a Gaussian distribution with \(\mu=0.5\) and \(\sigma=0.1\).
In [4]:
NDATA = 50000
# initialize data and randoms catalogs
data = RandomCatalog(NDATA, seed=42)
randoms = RandomCatalog(NDATA*10, seed=84)
# add the (ra, dec, z) columns
for s in [data, randoms]:
s['z'] = s.rng.normal(loc=0.5, scale=0.1, size=s.size)
s['ra'] = s.rng.uniform(low=110, high=260, size=s.size)
s['dec'] = s.rng.uniform(low=-3.6, high=60., size=s.size)
[ 000000.01 ] 0: 08-08 18:08 CatalogSource INFO total number of particles in RandomCatalog(size=50000, seed=42) = 50000
[ 000000.01 ] 0: 08-08 18:08 CatalogSource INFO total number of particles in RandomCatalog(size=500000, seed=84) = 500000
Adding the Cartesian Coordinates¶
Next, we add the Position column to both the “data” and “randoms” by
converting from sky coordinates to Cartesian coordinates, using the
helper function transform.SkyToCartesian. The redshift to comoving
distance transformation requires a cosmology instance, so we first
initialize our desired cosmology parameters.
In [5]:
# specify our cosmology
cosmo = cosmology.Cosmology(Om0=0.31, H0=70.0, flat=True)
# add Cartesian position column
data['Position'] = transform.SkyToCartesion(data['ra'], data['dec'], data['z'], cosmo=cosmo)
randoms['Position'] = transform.SkyToCartesion(randoms['ra'], randoms['dec'], randoms['z'], cosmo=cosmo)
Specifying the “data” \(n(z)\)¶
The ConvolvedFFTPower algorithm requires the number density as a
function of redshift for the “data” catalog. Here, we use the
RedshiftHistogram algorithm to compute the redshift histogram of the
“randoms” catalog, and then re-normalize the number density to that of
the “data” catalog.
In [6]:
# the sky fraction, used to compute volume in n(z)
FSKY = 0.15 # a made-up value
# compute n(z) from the randoms
zhist = RedshiftHistogram(randoms, FSKY, cosmo, redshift='z')
# re-normalize to the total size of the data catalog
alpha = 1.0 * data.csize / randoms.csize
# add n(z) from randoms to the FKP source
nofz = InterpolatedUnivariateSpline(zhist.bin_centers, alpha*zhist.nbar)
# plot
plt.plot(zhist.bin_centers, alpha*zhist.nbar)
plt.xlabel(r"$z$", fontsize=16)
plt.ylabel(r"$n(z)$ $[h^{3} \mathrm{Mpc}^{-3}]$", fontsize=16)
[ 000000.16 ] 0: 08-08 18:08 RedshiftHistogram INFO using Scott's rule to determine optimal binning; h = 4.40e-03, N_bins = 207
[ 000000.19 ] 0: 08-08 18:08 RedshiftHistogram INFO using cosmology {'H0': 70.0, 'Om0': 0.31, 'name': None, 'Ob0': 0.0486, 'Tcmb0': <Quantity 2.7255 K>, 'Neff': 3.04, 'm_nu': <Quantity [ 0., 0., 0.] eV>} to compute volume in units of (Mpc/h)^3
[ 000000.19 ] 0: 08-08 18:08 RedshiftHistogram INFO sky fraction used in volume calculation: 0.1500
Out[6]:
<matplotlib.text.Text at 0x1126ddd30>
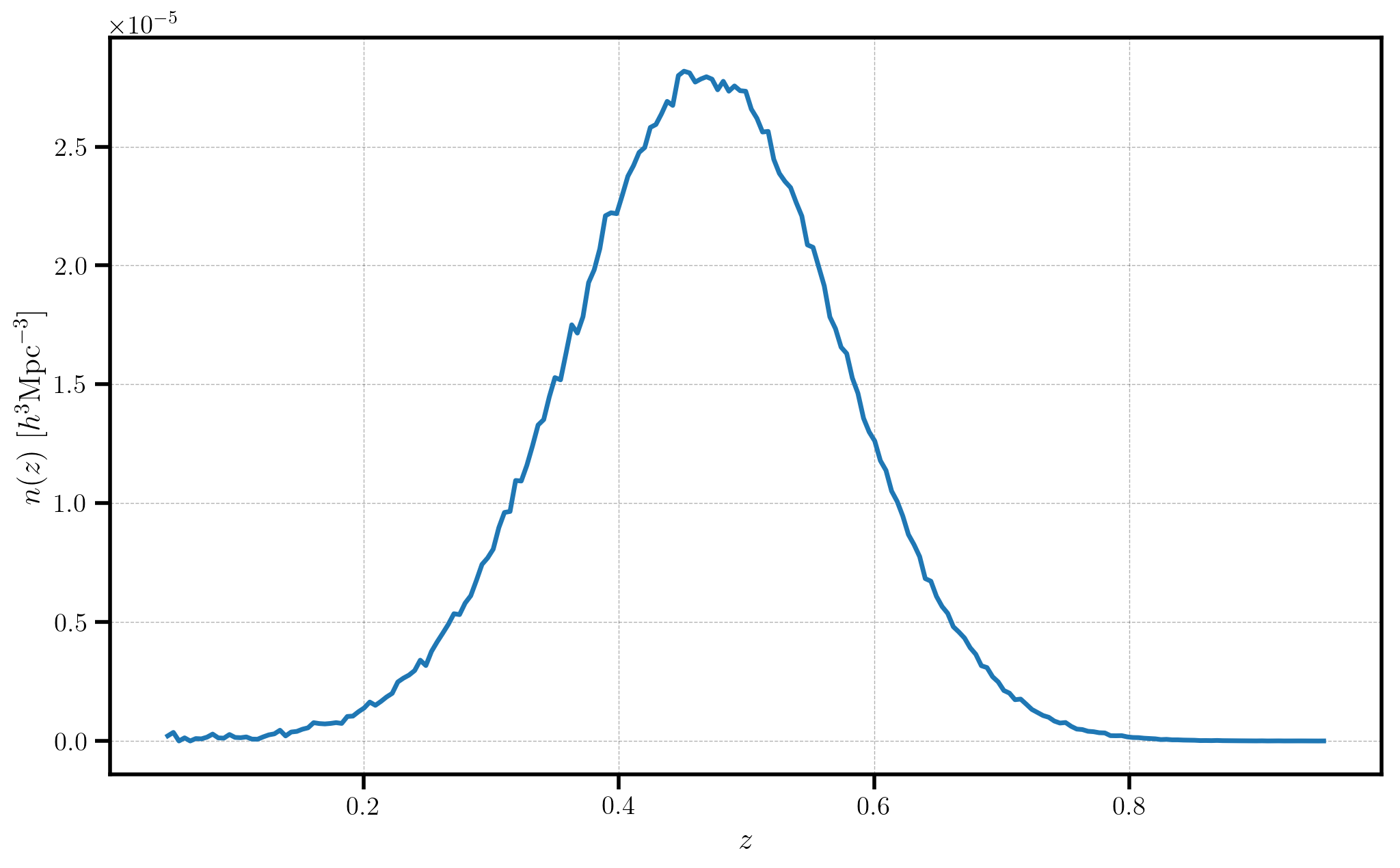
In this figure, note that the measured \(n(z)\) for the data closely resembles the input distribution we used: a Gaussian distribution with \(\mu=0.5\) and \(\sigma=0.1\).
Next, we initialize the FKPCatalog, which combines the “data” and
“randoms” catalogs into a single object. Columns are now available in
the FKPCatalog prefixed by either “data/” or “randoms/”.
In [7]:
# initialize the FKP source
fkp = FKPCatalog(data, randoms)
# print out the columns
print("columns in FKPCatalog = ", fkp.columns)
columns in FKPCatalog = ['data/FKPWeight', 'data/Position', 'data/Selection', 'data/TotalWeight', 'data/Value', 'data/Weight', 'data/dec', 'data/ra', 'data/z', 'randoms/FKPWeight', 'randoms/Position', 'randoms/Selection', 'randoms/TotalWeight', 'randoms/Value', 'randoms/Weight', 'randoms/dec', 'randoms/ra', 'randoms/z']
And we add the \(n(z)\) column to both the “data” and “randoms”, using the appropriate redshift column to compute the results.
In [8]:
# add the n(z) columns to the FKPCatalog
fkp['randoms/NZ'] = nofz(randoms['z'])
fkp['data/NZ'] = nofz(data['z'])
Adding FKP Weights¶
Here, we add a column FKPWeight that gives the appropriate FKP
weight for each catalog. The FKP weights are given by:
Here, we use a value of \(P_0 = 10^4 \ h^{-3} \mathrm{Mpc}^3\).
In [9]:
fkp['data/FKPWeight'] = 1.0 / (1 + fkp['data/NZ'] * 1e4)
fkp['randoms/FKPWeight'] = 1.0 / (1 + fkp['randoms/NZ'] * 1e4)
Adding Completeness Weights¶
The ConvolvedFFTPower algorithm also supports the use of
completeness weights, which weight the number density fields of the
“data” and “randoms” catalogs. Here, we add random weights to both
catalogs as the Weight column.
Completeness weights change the number density field such that the weighted number density field on the mesh is equal to \(n'(\mathbf{r}) = w_c(\mathbf{r}) n(\mathbf{r})\), where \(w_c\) represents the completeness weights.
In [10]:
fkp['data/Weight'] = numpy.random.random(size=data.size)
fkp['randoms/Weight'] = numpy.random.random(size=randoms.size)
Computing the Multipoles¶
To compute the multipoles, first we convert our FKPCatalog to a mesh
object, specifying the number of mesh cells per side, as well as the
names of the \(n(z)\) and weight columns.
If a Cartesian box size is not specified by the user, the size will be
computed from the maximum extent of the Position column
automatically.
In [11]:
mesh = fkp.to_mesh(Nmesh=256, nbar='NZ', comp_weight='Weight', fkp_weight='FKPWeight')
[ 000001.90 ] 0: 08-08 18:08 FKPCatalog INFO BoxSize = [ 2002. 3140. 2039.]
[ 000001.90 ] 0: 08-08 18:08 FKPCatalog INFO cartesian coordinate range: [-1984.36896194 -1127.47896845 -119.74859646] : [ -22.39806078 1950.19918467 1878.54836854]
[ 000001.90 ] 0: 08-08 18:08 FKPCatalog INFO BoxCenter = [-1003.38351136 411.36010811 879.39988604]
Now, we are able to compute the desired multipoles. Here, we compute the \(\ell=0,2,\) and \(4\) multipoles using a wavenumber spacing of \(k = 0.005\) \(h/\mathrm{Mpc}\). The maximum \(k\) value computed is set by the Nyquist frequency of the mesh, \(k_\mathrm{max} = k_\mathrm{Nyq} = \pi N_\mathrm{mesh} / L_\mathrm{box}\).
In [12]:
# compute the multipoles
r = ConvolvedFFTPower(mesh, poles=[0,2,4], dk=0.005, kmin=0.01)
[ 000001.93 ] 0: 08-08 18:08 ConvolvedFFTPower INFO using compensation function CompensateCICAliasing
[ 000003.16 ] 0: 08-08 18:08 FKPCatalogMesh INFO painting the 'data' species
[ 000003.16 ] 0: 08-08 18:08 CatalogSource INFO total number of particles in CatalogCopy(size=50000) = 50000
[ 000003.16 ] 0: 08-08 18:08 CatalogMesh INFO total number of particles in (CatalogCopy(size=50000) as CatalogMesh) = 50000
[ 000003.21 ] 0: 08-08 18:08 CatalogMesh INFO painted 50000 out of 50000 objects to mesh
[ 000003.21 ] 0: 08-08 18:08 CatalogMesh INFO mean particles per cell is 0.00125013
[ 000003.21 ] 0: 08-08 18:08 CatalogMesh INFO sum is 20973.6
[ 000003.21 ] 0: 08-08 18:08 CatalogMesh INFO normalized the convention to 1 + delta
[ 000003.22 ] 0: 08-08 18:08 FKPCatalogMesh INFO painting the 'randoms' species
[ 000003.23 ] 0: 08-08 18:08 CatalogSource INFO total number of particles in CatalogCopy(size=500000) = 500000
[ 000003.23 ] 0: 08-08 18:08 CatalogMesh INFO total number of particles in (CatalogCopy(size=500000) as CatalogMesh) = 500000
[ 000003.66 ] 0: 08-08 18:08 CatalogMesh INFO painted 500000 out of 500000 objects to mesh
[ 000003.66 ] 0: 08-08 18:08 CatalogMesh INFO mean particles per cell is -0.00124905
[ 000003.66 ] 0: 08-08 18:08 CatalogMesh INFO sum is 18.0156
[ 000003.66 ] 0: 08-08 18:08 CatalogMesh INFO normalized the convention to 1 + delta
[ 000003.84 ] 0: 08-08 18:08 ConvolvedFFTPower INFO cic painting done
[ 000004.24 ] 0: 08-08 18:08 ConvolvedFFTPower INFO ell = 0 done; 1 r2c completed
[ 000009.23 ] 0: 08-08 18:08 ConvolvedFFTPower INFO ell = 2 done; 5 r2c completed
[ 000017.65 ] 0: 08-08 18:09 ConvolvedFFTPower INFO ell = 4 done; 9 r2c completed
[ 000018.96 ] 0: 08-08 18:09 ConvolvedFFTPower INFO higher order multipoles computed in elapsed time 00:00:13.76
[ 000020.24 ] 0: 08-08 18:09 ConvolvedFFTPower INFO normalized power spectrum with `randoms.norm = 0.330767`
The meta-data computed during the calculation is stored in attrs
dictionary. See the
documentation for
more information.
In [13]:
for key in r.attrs:
print("%s = %s" % (key, str(r.attrs[key])))
poles = [0, 2, 4]
dk = 0.005
kmin = 0.01
use_fkp_weights = False
P0_FKP = None
BoxSize = [ 2002. 3140. 2039.]
BoxPad = [ 0.02 0.02 0.02]
BoxCenter = [-1003.38351136 411.36010811 879.39988604]
mesh.window = cic
mesh.interlaced = False
alpha = 0.100115376318
shotnoise = 39222.5631841
data.shotnoise = 35664.5480015
data.N = 50000
data.W = 25002.7957148
data.num_per_cell = 0.00125012579522
randoms.shotnoise = 3558.01518261
randoms.N = 500000
randoms.W = 249739.816542
randoms.num_per_cell = -0.00124905198259
data.norm = 0.329432188439
randoms.norm = 0.330767301835
The measured multipoles are stored in the poles attribute. Below, we
plot the monopole, quadrupole, and hexadecapole, making sure to subtract
out the shot noise value from the monopole.
In [14]:
poles = r.poles
for ell in [0, 2, 4]:
label = r'$\ell=%d$' % (ell)
P = poles['power_%d' %ell].real
if ell == 0: P = P - poles.attrs['shotnoise']
plt.plot(poles['k'], P, label=label)
# format the axes
plt.legend(loc=0)
plt.xlabel(r"$k$ [$h \ \mathrm{Mpc}^{-1}$]")
plt.ylabel(r"$P_\ell$ [$h^{-3} \mathrm{Mpc}^3$]")
plt.xlim(0.01, 0.3)
Out[14]:
(0.01, 0.3)
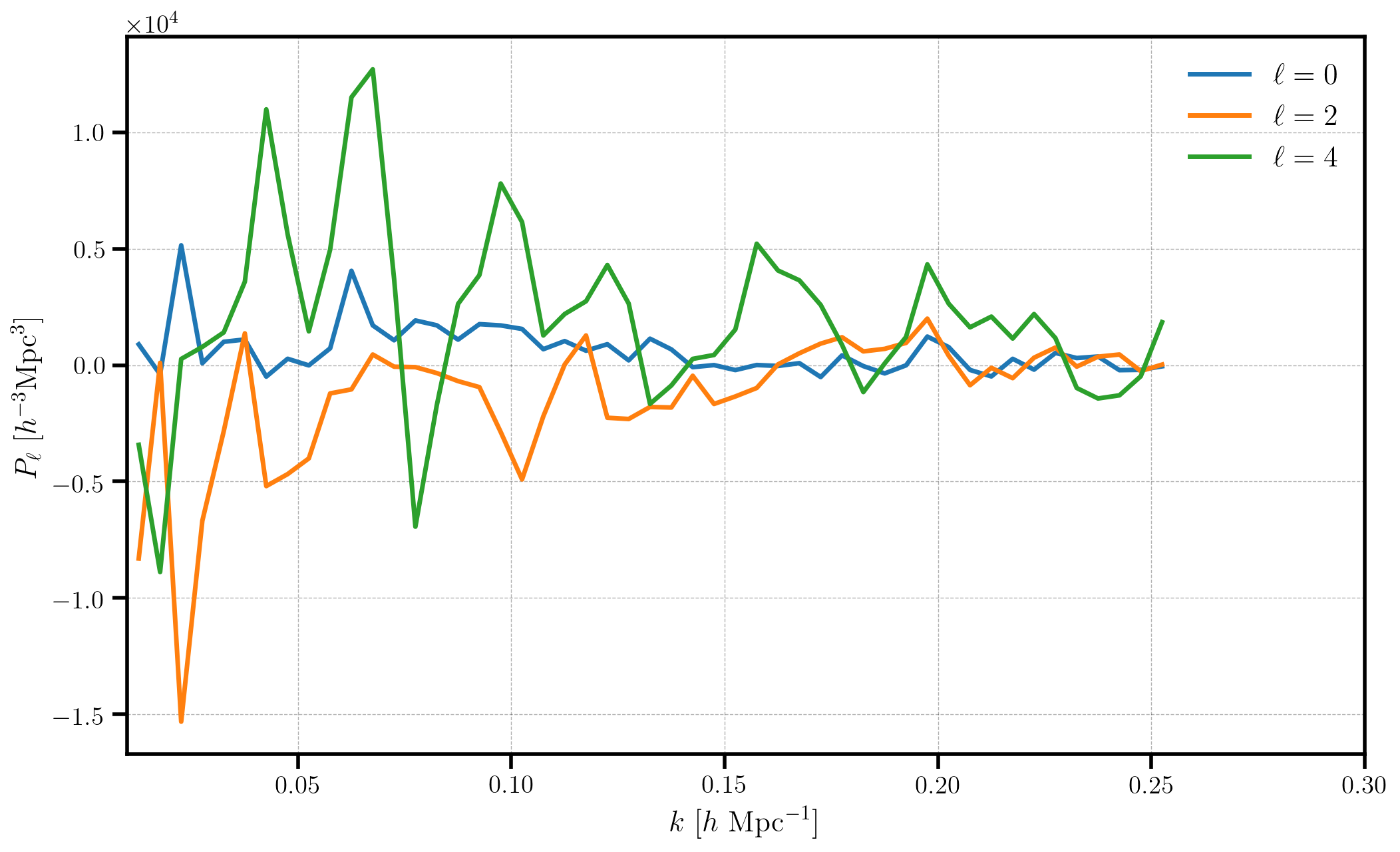
Note that, as expected, there is no measurably cosmological signal, since the input catalogs were simply uniformly distributed objects on the sky.